
|
LOVD v.2.0 - Leiden Open Variation Database
Online gene-centered collection and display of DNA variants
|
|
 |
Please note that LOVD2 is no longer supported. The last security release is from October 2020. Upgrade to LOVD3 to continue receiving updates. |
The LOVD system in short:
LOVD stands for Leiden Open (source) Variation Database.
LOVD's purpose : To provide a flexible, freely available tool for Gene-centered collection and display of DNA variants.
Mutalyzer
LOVD features integration with the Mutalyzer sequence variant nomenclature checker, allowing for direct nomenclature checking of sequence variants during the submission process.
 |
Are you interested in taking an LOVD course? Please send us a message and we'll keep you informed about the next LOVD course. |
LOVD's Popularity:
(Also see our public list of LOVD installations)
The number of IP adresses running an active LOVD (version 1.1.0-08 and up) installation, as measured until May, 2012. The total number of LOVD installations may be higher, since different installations at one server use one IP address. |
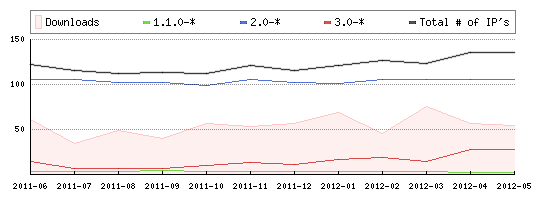 |
LOVD's statistics:
The total and unique number of variants contained in the single largest known LOVD install, as measured until May, 2012. |
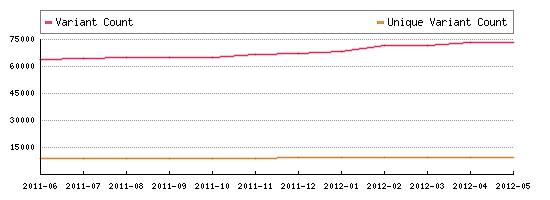 |
| Last modified 2019/02/21 15:22:59 CET |
When using or discussing LOVD please refer to:
Fokkema IF, Taschner PE, Schaafsma GC, Celli J, Laros JF, den Dunnen JT (2011). LOVD v.2.0: the next generation in gene variant databases.
Hum Mutat. 2011 May;32(5):557-63.
|
|


