|
|
LOVD ManualManager handbookIn LOVD, there are three levels of registered users (not submitters). The Database manager, or DB manager, has full access to every functionality available in LOVD. There can be only one DB manager.A manager has access to almost everything the DB manager has access to. However, the manager is not able to create new managers or to edit existing managers. Also, a manager cannot uninstall LOVD. A curator has only access to genes the curator has been authorized for. All functionality in this chapter is available from the level of manager. Some functionality is limited to the DB manager. If so, this is clearly indicated. You can log in by pointing your browser to the config directory. System wide setup
System settingsGeneral system settingsThese settings are used to somewhat update the LOVD behaviour to your needs.Database location: This value will be viewed on the gene homepages and will also be used in certain emails LOVD sends. It's default value is autodetected. LOVD Email address: This value will be used by LOVD to send emails with, because emails without a 'From' address may be blocked by some SPAM filters. Allow public to download your gene tables: Select whether or not the public is allowed to download your gene tables from the gene homepages. This option is unchecked by default. Refresh variant list after modification: Select whether or not you want the active list of variants to refresh when you have just added, edited, duplicated or deleted an entry from that list. This option is checked by default. Use cookies for user authentication: Select whether or not you want to use cookies for user authentication. Cookies are small text-files sent back and forth to the user and LOVD to identify the user with the system. Leaving this option set is highly recommended, as disabling this may allow an attacker to hijack a LOVD user account. Legend settingsThese settings are used to create a more detailed legend. Also, the lists of phenotypes and techniques are used to create selectionlists on the variant submission form.List of possible phenotypes for this LOVD installation: Specify a list of phenotypes that are used in this LOVD installation. This list will be put on the full legend and will allow a submitter to select a phenotype from the list. If no phenotypes are specified, the submitter will need to fill in the phenotype in a common text field. Format: one phenotype per line; Abbreviation = Phenotype name. Example: "DMD = Duchenne muscular dystrophy". URL explaining the phenotypes: Specify an url with more information about the phenotypes mentioned in the list above. For instance, a website with clinical features. Fill in the complete URL, including the "http://". List of available detection techniques: Specify a list of detection techniques appropriate for the genes configured in this LOVD installation. By default, a full set of detection techniques have been put in this list already. This list will be put on the full legend and will allow a submitter to select the used detections technique(s) from the list. If no techniques are specified, the submitter will need to fill in the technique in a common text field. Format: one technique per line; Abbreviation = Technique name. Example: "DHPLC = Denaturing High-Performance Liquid Chromatography". URL explaining the techniques: Specify an url with more information about the detection techniques mentioned in the list above. For instance, a website with available primer sequences. Fill in the complete URL, including the "http://". Custom columnsThe custom columns added to LOVD will appear directly after the Remarks column. You can change the order of the custom columns, but you can not place them before the Remarks column or after the Patient ID field. Creating new columns
Title: Type in a title that will show on top of the column and on the forms. Unique abbreviation code: Choose an unique abbreviation for the column that will be used to identify it in the database. This abbreviation must start with a letter, be 3-20 characters long, and may contain lowercase letters, numbers and underscores (_) only. Column type: Select the type of column: numeric, text (up to 255 characters), or large text. Maximum length of column values: Type in the maximum value length for the column. Public: Select whether or not you want the column to be visible to the public. Editing existing columnsTo edit a column, click on the title. You will get a form similar to the form used to create custom columns. Deleting existing columnsClick on the 'Edit Cols' button in the Setup area menu. Then click on the small trashbin directly at the right-hand side of the column you wish to delete. You will be prompted to fill in your password, after which the column will be removed from all gene tables.Custom links
References are used to pass on information necessary for building the link, e.g. a PubMed ID. The references need to be specified in both the link pattern and the replacement text, and are indicated by numbers surrounded by square brackets, like '[1]' or '[2]'.
Creating new linksTitle: Fill in a short title that will make curators recognize the target of the link. Pattern: Choose a pattern that curators need to use for LOVD to recognize the custom link. This pattern should always start with a '{' and end with a '}'. Between these braces only letters, numbers, spaces, some special characters (:;,_-) and references ([1] to [99]) may be allowed. The custom link pattern must be between 3-20 characters long. Replacement text: Fill in the (HTML enabled) text that should replace the entire pattern. Active for columns: Select for which columns you want this custom link to be activated. Replacement active: Select whether or not you want this custom link to be activated. If 'No' is selected, found patterns will not be replaced. Editing existing linksDeleting existing linksClick on the 'Edit Links' button in the Setup area menu. Then click on the small trashbin directly at the right-hand side of the link you wish to delete. You will be prompted if you are sure to delete the selected link.
Gene specific setupWhen creating new genes and editing existing ones, you can set some gene-specific settings to customize the LOVD public area a bit. For example, you can add links, remarks and OMIM disease ids to the gene homepage. Please note that since curators cannot create new genes or edit existing ones, these options are only available to managers.If you wish to personalize your LOVD installation somewhat, you can also add headers and footers to the public area. To do so, create text files in the include directory and name them files 'GENESYMBOL_top.txt' or 'GENESYMBOL_bot.txt' (for example, 'DMD_top.txt'). The header and footer will, if present, be viewed on the gene homepage, variant tables, search forms and search results. The files are HTML enabled. Creating new genes
The form used to create a new gene database also allows you to set some gene-specific settings. The top part, 'General options', is mostly mandatory to fill in. Only the 'Date of creation' is optional. The entire bottom part of the form, 'Optional options', is optional. LOVD will create a gene homepage for you, from which the allelic variants can be accessed and external data sources can be linked. The 'Optional options' are mainly to adapt this gene home page to your needs. General optionsFull gene name:Fill in the full gene name, such as 'Duchenne Muscular Dystrophy'. Gene Symbol: Fill in the official gene symbol of the gene. If you wish to create multiple gene databases for only one gene (for instance, for different types of allelic variants), use an underscore after the gene symbol, followed by a short unique string of characters. For example, 'DMD_point' for point mutations in the DMD gene and 'DMD_deldup' for large multi-exon deletions or duplications in the DMD gene. Chromosome Location: Fill in the chromosomal location of the gene. Date of creation: The date creation is put on the gene homepage and would normally be the date the gene was created in LOVD. However, you may have published your data before somewhere and would want to use the date of first publication. In that case, you can fill in that date in this field. Format: YYYY-MM-DD or YYYY/MM/DD. Example: 1997-01-10. Optional optionsHomepage:If you have set up an own web page about the gene in question, you can fill in the full address (URL) here, including the 'http://'. External links: If you have other links that you want included in the gene homepage, specify them here. You can use the full address (URL) including the 'http://' or use a format of 'Description of page <URL>'. Multiple links can be separated by semicolons (;). Hide second allele in tables: Select whether or not you want the second allele field to be hidden. If the gene is on the X-chromosome, you may consider the second allele irrelevant. Entrez Gene (Locuslink) ID: Fill in the Entrez Gene (formerly known as Locuslink) ID here to add a link to Entrez Gene from the LOVD gene homepage. OMIM Gene ID: Fill in the OMIM ID here to add a link to OMIM from the LOVD gene homepage. OMIM Disease IDs: Fill in one or more OMIM Disease ID's here to provide links to those OMIM pages from the LOVD gene homepage. Format: OMIM_ID Disease_name; OMIM_ID Disease_name [;...] Example: 310200 DMD;300376 BMD. HGMD ID: Fill in the HGMD ID, to create a link to HGMD from the LOVD gene homepage. GDB ID: Fill in the GDB ID (usually the same as the HGMD ID), to create a link to GDB from the LOVD gene homepage. Notes for the indexpage: There is some room for notes on the LOVD gene homepage. These notes are HTML enabled. Notes for the searchpage: In case you use any special symbols or specific formats in your listings that might need some explanation, text in this field is printed below all allelic variant listings, just above the legend. These notes are HTML enabled. Reference for exon numbering: If available, specify the reference that first described the structure of the gene. This will be put in the legend as 'Exon numbering according to ...'. Has a reference sequence: Select if a reference sequence exists, and if yes, what type. Please note that the LOVD Reference Sequence parser is available to create cDNA reference sequences. Reference sequence URL: If a reference sequence is available, full in the address (URL) of the reference sequence of this gene. If the reference sequence was created using the LOVD Reference Sequence parser, the location of the reference sequence is printed below this field. Editing existing genesThen click the 'Edit Db' button. You will get a form similar to the form used to create new gene databases. Deleting existing genesThen click the 'Drop Db' icon. You will need to fill in your password for authentication. After submitting the form, you are once more asked to provide your password to completely remove the gene and it's variants from the database.
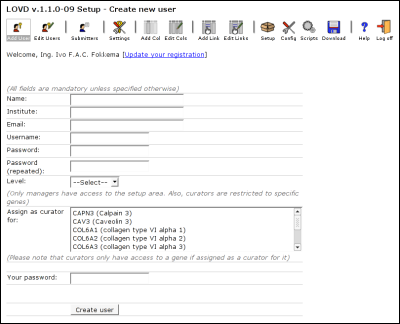
User managementThe first users within LOVD is always the Database manager. This account is created at installation of LOVD. It is then the task of the DB manager to create other users (managers and curators) in the system, if desired. Managers are only able to create or edit curators in LOVD.User levelsCreating, editing and deleting users is only possible with users with a lower level than you. In other words, when you are a manager, you can create, edit and delete curators only. As the DB manager, you can create, edit and delete managers and curators.Database (DB) managerThe first registered LOVD user, created at installation. There is only one DB manager and there is no restriction to the access of the DB manager. Also, the DB manager is the only user that can uninstall LOVD, provided the uninstall lock has not been set at LOVD installation.The DB manager may, depending on the config.ini settings, receive copies of submitter registrations and variant submissions. This is on by default, but can be turned off by setting the 'copy' directive, at the bottom of the config.ini file, to 'Off'. Especially when the DB manager has no further role within LOVD, for instance when a computer technician is asked to install LOVD instead of a researcher or with LOVD installations with large quantities of gene databases and/or registered submitters, the reports serve no further purpose. ManagerA manager has access to almost everything the DB manager has access to. However, the manager is not able to create new managers or to edit existing managers. Also, a manager cannot uninstall LOVD even if the uninstall lock is not in place.A manager is usually created by the DB manager to assist the DB manager or for taking over the customisation of LOVD for the curators. CuratorA curator only has access to the gene databases he or she is appointed to by a (DB) manager. A manager (or DB manager) can also act as a curator, so the curator user level is not strictly required. It is, however, very useful for allowing people access to certain genes while access to LOVD settings and such is restricted. If you make no use of the curator user level, make sure you select a manager as gene curator by selecting the relevant genes in the list "Curator for" when creating or editing the targeted manager.Creating new users
Name: The user's name. Institute: Fill in the user's institute. Email: The user's email. If this user is to be a gene's curator, this email address will show up on that gene's homepage and the user will receive submissions sent for that gene on this email address. Username: Pick a username for the user. It has to be unique; e.g. when you fill in a username that is already taken by another user, you will have to pick another one. Password: Fill in the user's password. Password (repeated): Retype the user's password. This is to make sure the password is put in the database correctly, it is less likely for you to make a typo twice. Level: Select the user's level. If you are the DB manager, this list will contain 'Manager' and 'Curator'. If you are a manager, this list will only have the 'Curator' option. Curator for: If the user is to be a curator, select the gene(s) the user should be allowed access to. If you forget this, the curator can't access LOVD, since there are no genes open to this user. If the user is a manager, selecting genes will not restrict the manager access to those genes only, but it will select the user as a curator for these genes. This may be useful if you have not created any curators in the system, but are curating the few genes in your installation yourself. Your password: Fill in your own password for authorization. This makes sure that if you would leave your computer while logged into LOVD, no one can create a new user within LOVD without knowing your password. Editing existing usersPlease note that the "New password" fields are optional when editing a user, you only have to fill them in when changing the password for the user. Deleting existing usersClick on the 'Edit Users' button in the Setup area menu. Then click on the small trashbin directly at the right-hand side of the user you wish to delete. You will be required to fill in your password to delete the specified user.If the deleted user was a curator for a gene that would now be left without any curator, you will automatically take over that gene. Even as a manager, your name will then be listed as the gene's curator on the LOVD gene homepage. This is done by selecting the gene in your 'Curator for' list. In order to prevent this from happening, make sure the gene has at least one other curator by selecting the gene in the 'Curator for' list of a different user. For more information on what happens when a gene is left without a curator, see below. Assigning curators to specific genes
Please note that a curator for a gene is not necessarily a user with the 'Curator' user level. A manager, or even the DB manager, can be appointed as a gene curator. This is done so that LOVD installations with for instance but one gene, can be both configured and curated by one user with manager clearance. If a gene is left without a curator, as is the case after a gene database has been created, the LOVD gene homepage does not show a name or email address of the responsible curator. When a submission is done for this gene, no curator will be able to receive a copy of the submission data. To make sure the submission will not go unnoticed, the DB manager will receive a copy of the submission data, in stead of a curator. This is done regardless of the 'copy' setting in the config.ini file. If multiple users are assigned curator for a gene, all users will receive the submission email. Curator level users will not be able to log into LOVD if no genes have been assigned to them. To assign genes to curators or vice versa, click on the 'Edit Users' button in the Setup area menu. You will see a list of the users within LOVD. To assign a gene to a user, click on the name of that user. You will get a form similar to the form used to create new users. Select the gene(s) you want the user to be curator for in the list 'Curator for', fill in your password in the 'Your password' field and click the 'Edit user' button. Curator level users will not be able to switch to any gene other than the genes in their allowed list. Managers and the DB manager will not be restricted access to any gene, but the list may be used to assign a manager as curator for a gene. Uninstalling LOVD (DB manager only)Only the DB manager can uninstall LOVD, provided the uninstall lock has not been set at install of LOVD. The uninstall lock is set within the MySQL database and can only be removed by directly accessing MySQL. Uninstalling LOVD can also be done by using MySQL directly.Go to the Setup area and click on the uninstall link at the bottom. See the figure at the start of this chapter. You will need to fill in the DB manager's password twice to proceed.
|